sceasy转h5ad到rds之后基因名字消失的解决方案
|Word Count:182|Reading Time:1mins|Post Views:
最近在做各种各样的物种的单细胞数据清洗,在清洗到某个数据的时候,用sceasy转格式从h5ad到rds,发现了一个问题,基因名字消失了。
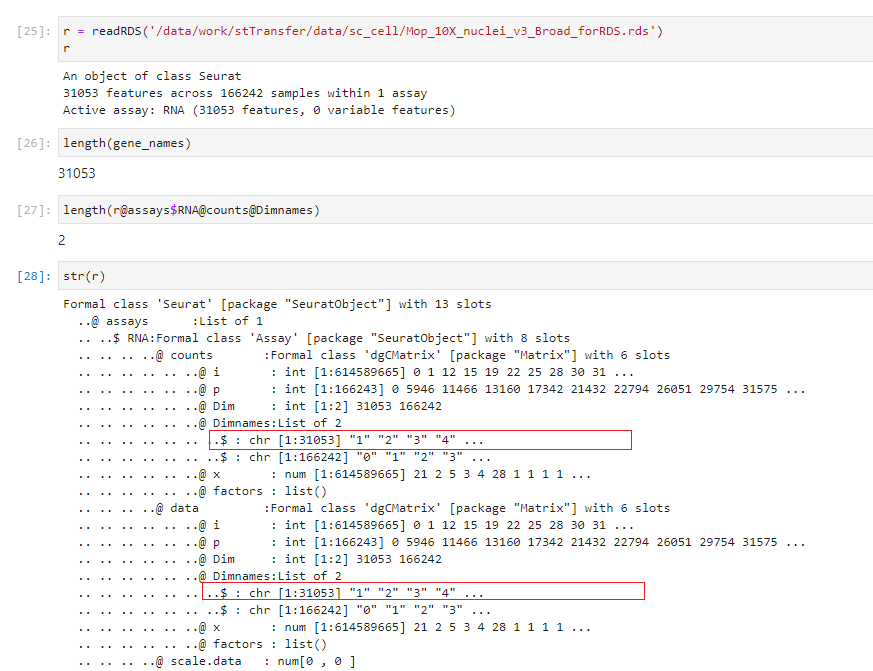
没有再去看sceasy的源码,从源码去解决问题,只能是把原先的基因名字进行迁移。
1
2
|
adata.var.to_csv('Mop_10X_nuclei_v3_Broad_forRDS.csv')
|
接着是rds的基因名字的赋予
1
2
3
4
5
6
7
8
9
10
11
12
|
df <- read.table('/data/work/stTransfer/data/sc_cell/Mop_10X_nuclei_v3_Broad_forRDS.csv', header = FALSE)
r = readRDS('/data/work/stTransfer/data/sc_cell/Mop_10X_nuclei_v3_Broad_forRDS.rds')
counts= r@assays$RNA@counts
meta = r@meta.data
counts <- as.matrix(counts)
counts = data.frame(counts)
rownames(counts) = gene_names
data_m = CreateSeuratObject(counts = counts, metadata = meta)
saveRDS(data_m, file = "your_file_name.rds")
|
