计算某个区域中细胞的复杂性 | Word Count: 944 | Reading Time: 4mins | Post Views:
在某系空间组图谱文章中,我们经常会看到一种图片,是去计算某个区域中细胞的复杂性,我们以今年在
Cell
上发表的一篇文章为例,来看看如何计算某个区域中细胞的复杂性。文章名字为Single-cell
spatial transcriptome reveals cell-type organization in the macaque
cortex,文章链接为:https://linkinghub.elsevier.com/retrieve/pii/S0092867423006797
,图二中有这么一张图片:
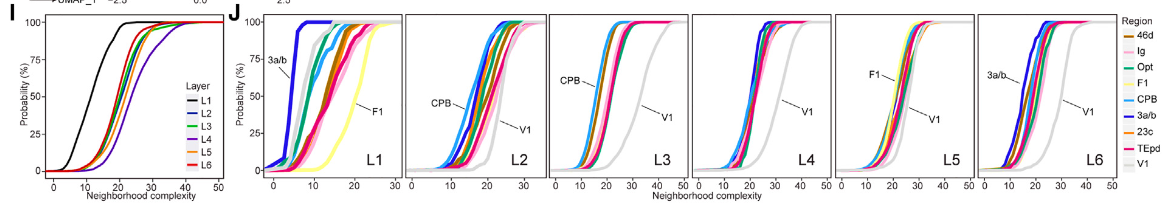
I图的意思是不同的 Layer
中的每个细胞的复杂度分布,每个细胞的复杂的计算为,以每个细胞为圆心,200个像素为半径,画一个圆圈,计算这个圆圈中和这个细胞的类型不同的细胞类型个数。根据这个理解,我们便是可以写程序了。
1 2 3 4 5 6 7 8 9 10 import anndata as adimport numpy as npimport pandas as pdimport scanpy as scfrom matplotlib.ticker import FuncFormatterimport seaborn as snsimport numpy as npfrom collections import Counterimport matplotlib.pyplot as plt
这里我写了三个函数,三个函数的计算速度是一个比一个快,但是易理解程度是一个比一个难
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 def calculate_neighborhood_complexity (cell_coordinates, cluster_labels, radius ): neighborhood_complexities = [] for central_cell, central_cluster_label in zip (cell_coordinates, cluster_labels): complexity = 0 for cell, cluster_label in zip (cell_coordinates, cluster_labels): distance = np.linalg.norm(cell - central_cell) if 0 < distance <= radius and cluster_label != central_cluster_label: complexity += 1 neighborhood_complexities.append(complexity) complexity_counts = Counter(neighborhood_complexities) total_cells = len (neighborhood_complexities) complexity_probabilities = { complexity: count / total_cells for complexity, count in complexity_counts.items() } return np.array(neighborhood_complexities), complexity_probabilities def calculate_neighborhood_complexity (cell_coordinates, cluster_labels, radius ): unique_labels, label_indices = np.unique(cluster_labels, return_inverse=True ) complexity_counts = np.zeros(len (cell_coordinates), dtype=int ) for i, central_cell in enumerate (cell_coordinates): distances = np.linalg.norm(cell_coordinates - central_cell, axis=1 ) mask = np.logical_and(distances > 0 , distances <= radius) previous_complexities = set () for cell_idx, cluster_idx in enumerate (label_indices[mask]): if cluster_idx not in previous_complexities: complexity_counts[i] += 1 previous_complexities.add(cluster_idx) total_cells = len (cell_coordinates) complexity_probabilities = np.bincount(complexity_counts) / total_cells return complexity_counts, complexity_probabilities from scipy.spatial import cKDTreedef calculate_neighborhood_complexity (cell_coordinates, cluster_labels, radius ): unique_labels, label_indices = np.unique(cluster_labels, return_inverse=True ) complexity_counts = np.zeros(len (cell_coordinates), dtype=int ) kdtree = cKDTree(cell_coordinates) for i, central_cell in enumerate (cell_coordinates): neighbor_indices = kdtree.query_ball_point(central_cell, radius) previous_complexities = set () for cell_idx, cluster_idx in zip (neighbor_indices, label_indices[neighbor_indices]): if cluster_idx not in previous_complexities: complexity_counts[i] += 1 previous_complexities.add(cluster_idx) total_cells = len (cell_coordinates) complexity_probabilities = np.bincount(complexity_counts) / total_cells return complexity_counts, complexity_probabilities
那么应该如何是用,以及如何绘制图片呢?
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 cell_coordinates_list = [ adata[adata.obs['region' ]=='layer1' ].obsm['spatial' ], adata[adata.obs['region' ]=='layer2' ].obsm['spatial' ], adata[adata.obs['region' ]=='layer3' ].obsm['spatial' ], adata[adata.obs['region' ]=='layer4' ].obsm['spatial' ], ] cluster_labels_list = [ adata[adata.obs['region' ]=='layer1' ].obs['celltype' ].tolist(), adata[adata.obs['region' ]=='layer2' ].obs['celltype' ].tolist(), adata[adata.obs['region' ]=='layer3' ].obs['celltype' ].tolist(), adata[adata.obs['region' ]=='layer4' ].obs['celltype' ].tolist(), ] name = ['layer1' , 'layer2' , 'layer3' , 'layer4' ] radius_list = [200 , 200 , 200 , 200 ] fig, ax = plt.subplots(figsize=(6 , 6 )) for i, (cell_coordinates, cluster_labels, radius) in enumerate (zip (cell_coordinates_list, cluster_labels_list, radius_list)): complexities, complexity_probabilities = calculate_neighborhood_complexity(cell_coordinates, cluster_labels, radius) sns.kdeplot(complexities, cumulative=True , fill=False , ax=ax, label=name[i]) ax.set_xlabel('Neighborhood Complexity' ) ax.set_ylabel('Probability (%)' ) ax.set_title('Brain Neighborhood Complexity Distribution' ) ax.legend(loc='lower right' ) ax.yaxis.set_major_formatter(FuncFormatter(lambda x, _: f'{x*100 :.0 f} ' )) plt.tight_layout() plt.savefig('Brain_Neighborhood_Complexity_Distribution.pdf' ) plt.show()
画出来的结果便是 Cell 的图片的样子了