计算某个区域中细胞的复杂性(2) | Word Count: 776 | Reading Time: 3mins | Post Views:
上一篇帖子提到了猴脑文章中某个区域中细胞复杂性的计算 ,后面又看到一张图,才是发现自己的计算少了一些东西,这里补充一下。
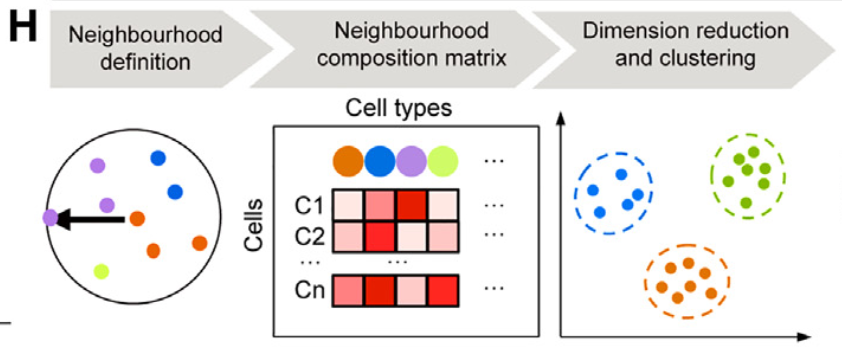
在原文的图四中还有一个图片,讲述了较为详细的计算过程,如下图所示:
首先是复杂性的获得,我的上一篇相关的博客已经讲了,但是那篇博客的问题也是在于此,我是直接获得了复杂性,把其他的信息都丢掉了,KD树的缺陷也在于此,于是不得已在此进行重构:
1 2 3 4 5 6 7 8 9 10 11 import numpy as npfrom scipy.spatial.distance import cdistimport anndata as adimport pandas as pdimport matplotlib.pyplot as pltfrom collections import Counterimport seaborn as snsfrom matplotlib.ticker import FuncFormatterfrom matplotlib.colors import ListedColormapfrom operator import itemgetter
然后是计算函数的定义:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 def calculate_neighborhood_complexity (coordinates, cell_types ): ''' coordinates: 细胞的坐标 list cell_types: 细胞的类型 list ''' num_cells = len (coordinates) unique_cell_types = np.unique(cell_types) num_cell_types = len (unique_cell_types) cell_type_indices = {cell_type: i for i, cell_type in enumerate (unique_cell_types)} complexity_matrix = np.zeros((num_cells, num_cell_types), dtype=int ) for i in range (num_cells): center_cell_type = cell_types[i] center_coord = coordinates[i] distances = cdist([center_coord], coordinates)[0 ] neighborhood_indices = np.where(distances <= 200 )[0 ] neighborhood_cell_types = cell_types[neighborhood_indices] unique_clusters, cluster_counts = np.unique(neighborhood_cell_types, return_counts=True ) for cell_type, count in zip (unique_clusters, cluster_counts): column_index = cell_type_indices[cell_type] complexity_matrix[i, column_index] += count complexity_matrix = pd.DataFrame(complexity_matrix, columns=unique_cell_types, ) complexity = (complexity_matrix != 0 ).sum (axis=1 ) complexity_matrix['complexity' ] = complexity complexity_matrix['cell_types' ] = cell_types return complexity_matrix
如何使用呢?如下所示:
1 2 3 4 5 6 7 adata = ad.read('/data/cellbin_transAnno.h5ad' ) coordinates = adata_temp.obsm['spatial' ] cell_types = np.array(adata_temp.obs['celltype' ].tolist()) complexity_matrix = calculate_neighborhood_complexity(coordinates, cell_types) complexity_matrix.to_csv('/data/complexity_matrix.csv' )
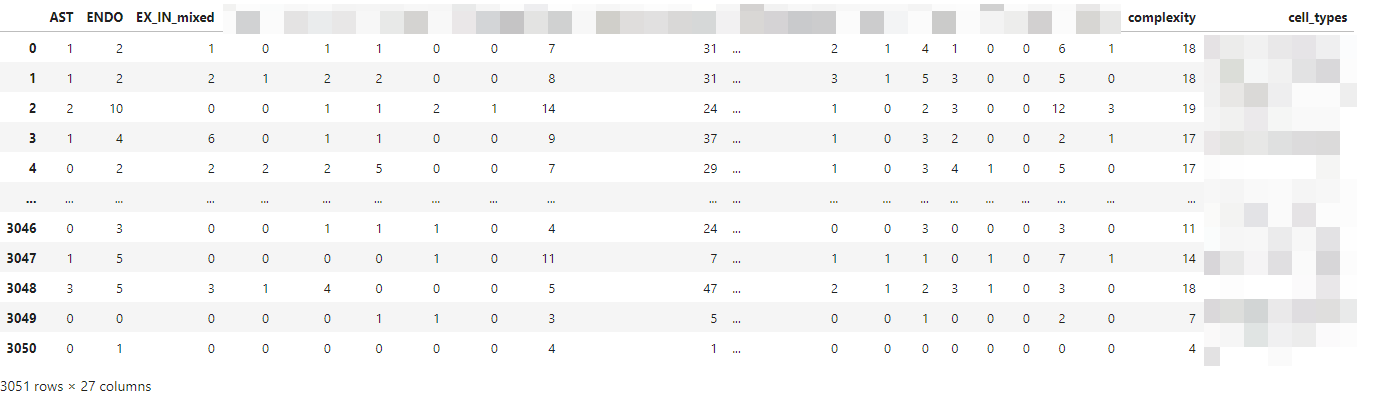
这里的 complexity_matrix 是一个 pandas 的 DataFrame,不会用 python
画图的友友可以保存为 csv 文件,然后用 R 语言画图,本人不是很会使用 R
语言,所以就不提供 R 语言的代码了,dataframe
概览如下所示,每一行为一个细胞,每一列为一个细胞类型,最后两列为每个细胞的复杂性和细胞类型,中间的值为
distances 在 200 之内的 某个细胞类型的数量。
本人绘图的代码如下所示
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 adata_temp = adata coordinates = adata_temp.obsm['spatial' ] cell_types = np.array(adata_temp.obs['celltype' ].tolist()) complexity_matrix = calculate_neighborhood_complexity(coordinates, cell_types) ex = ['X1' , 'X2' , 'X3' ] complexity_matrix = complexity_matrix[complexity_matrix['cell_types' ].isin(ex)] fig, ax = plt.subplots(figsize=(9 , 6 )) for j in ex: temp = complexity_matrix[complexity_matrix['cell_types' ] == j] if len (temp)>0 : neighborhood_complexities = temp['complexity' ].tolist() sns.kdeplot(neighborhood_complexities, cumulative=True , fill=False , ax=ax, label=j, color = celltypes_colors[j]) else : continue ax.set_xlabel('Neighborhood Complexity' ) ax.set_ylabel('Probability (%)' ) ax.set_title('ALL Neighborhood Complexity Distribution' ) ax.legend(loc='upper left' , bbox_to_anchor=(1.02 , 1 )) ax.yaxis.set_major_formatter(FuncFormatter(lambda x, _: f'{x*100 :.0 f} ' )) ax.xaxis.set_major_locator(plt.MaxNLocator(integer=True )) plt.tight_layout() plt.savefig('Neighborhood_Complexity_Distribution.pdf' ) plt.close()
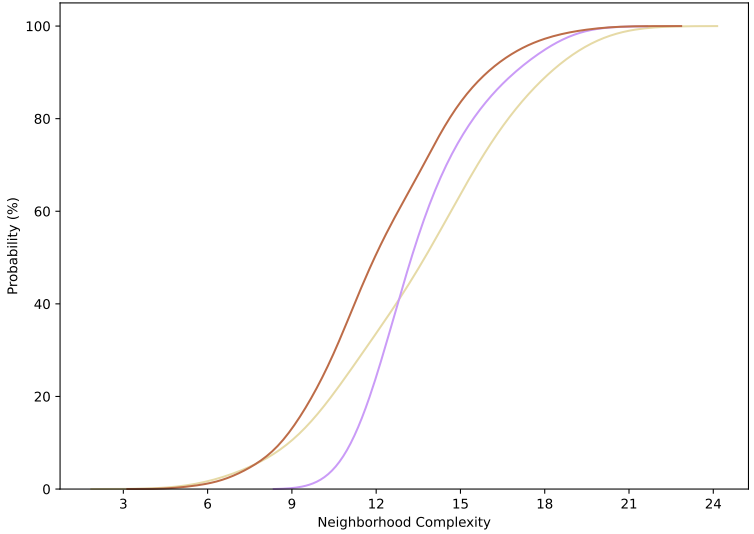
绘图结果如下所示:
以上